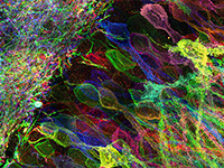
Tools for mapping the molecules and structure of the brain
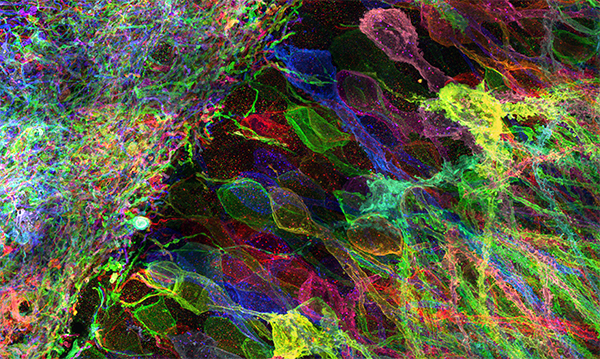
Biological systems such as brain circuits are complex structures made out of nanoscale building blocks such as proteins, RNAs, DNA, lipids, sugars, and other biomolecules, which take a great many forms, and are often organized with nanoscale precision. This presents a fundamental tension in biology – to understand a biological system like a brain circuit, you might need to map a large diversity of nanoscale building blocks, with great precision, across 3D spatial expanses. We are developing a suite of tools that enable the mapping of the location and identity of the molecular building blocks of biological systems such as the brain, aiming to map out the architecture of such systems with enough precision to understand how the structures of biological systems lead to function and dysfunction. One of the technologies we are developing, expansion microscopy (ExM), enables specimens to be imaged with nanoscale precision on ordinary microscopes, by physically expanding preserved biological systems (in contrast to all previous microscopy technologies, that magnify images of specimens via lenses). We are working to improve expansion microscopy further, improving resolution and multiplexing capability, and are working, often in interdisciplinary collaborations, on a suite of new labeling and analysis techniques that exploit the biochemical freedom enabled by the expanded state. We are working, often in interdisciplinary collaborations, for example, on ways to perform in situ RNA and DNA sequencing in intact biological systems, or to visualize and identify individual protein molecules within intact cells and tissues. We share all such tools as freely as possible, aiming to democratize multiplexed, nanoresolution imaging by making it as powerful and inexpensive as possible. We are also applying expansion microscopy to the scalable mapping of biological systems, such as brain circuits. Such brain circuit maps may be detailed enough someday to enable detailed computer simulations of brain functions, starting with small organisms such as C. elegans and zebrafish, and scaling up to the mouse and human brain. Finally, we are extending and applying such tools to the early detection and understanding of complex diseases such as cancers and immune diseases, and to the analysis of brain aging and aging more broadly.
Publications
Combinatorial protein barcodes enable self-correcting neuron tracing with nanoscale molecular context
bioRxiv | 2025Park SY*, Sheridan A*, An B, Jarvis E, Lyudchik J, Patton W, Axup JY, Chan SW, Damstra HGJ, Leible D, Leung KS, Magno CA, Meeran A, Michalska JM, Rieger F, Wang C, Wu M, Church GM, Funke J, Huffman T, Leeper KGC**, Truckenbrodt S**, Winnubst J**, Kornfeld JMR, Boyden ES+, Rodriques SG***, Payne AC+*** (2025) Combinatorial protein barcodes enable self-correcting neuron tracing with nanoscale molecular context, bioRxiv 2025.09.26.678648, doi: 10.1101/2025.09.26.678648. (*, contributed equally; **, contributed equally; ***, contributed equally; +, co-corresponding)
Expansion in situ genome sequencing links nuclear abnormalities to aberrant chromatin regulation
Science | 2025Labade AS, Chiang ZD, Comenho C, Reginato PL, Payne AC, Earl AS, Shrestha R, Duarte FM, Habibi E, Zhang R, Church GM, Boyden ES, Chen F, Buenrostro JD (2025) Expansion in situ genome sequencing links nuclear abnormalities to aberrant chromatin regulation, Science 389(6758):eadt2781.
Nanoscale Resolution Imaging of Whole Mouse Embryos Using Expansion Microscopy
ACS Nano | 2025Sim J, Park CE, Cho I, Min K, Eom M, Han S, Jeon H, Cho ES, Lee Y, Yun YH, Lee S, Cheon DH, Kim J, Kim M, Cho HJ, Park JW, Kumar A, Chong Y, Kang JS, Piatkevich KD, Jung EE, Kang DS, Kwon SK, Kim J, Yoon KJ, Lee JS, Kim CH, Choi M, Kim JW, Song MR, Choi HJ, Boyden ES, Yoon YG, Chang JB (2025) Nanoscale Resolution Imaging of Whole Mouse Embryos Using Expansion Microscopy, ACS Nano, doi:10.1021/acsnano.4c14791. Online ahead of print.
Dense, continuous membrane labeling and expansion microscopy visualization of ultrastructure in tissues
Nature Communications | 2025Shin TW, Wang H*, Zhang C*, An B, Lu Y, Zhang E, Lu X, Karagiannis ED, Kang JS, Emenari A, Symvoulidis P, Asano S, Lin L, Costa EK; IMAXT Grand Challenge Consortium; Marblestone AH, Kasthuri N, Tsai LH, Boyden ES (2025) Dense, continuous membrane labeling and expansion microscopy visualization of ultrastructure in tissues, Nature Communications 16(1):1579. (*, contributed equally)
High-throughput expansion microscopy enables scalable super-resolution imaging
eLife | 2024Day JH, Della Santina CM, Maretich P, Auld AL, Schnieder KK, Shin T, Boyden ES, Boyer LA (2024) High-throughput expansion microscopy enables scalable super-resolution imaging, Elife 13:RP96025.
Multiplexed expansion revealing for imaging multiprotein nanostructures in healthy and diseased brain
Nature Communications | 2024Kang J*, Schroeder ME*, Lee Y, Kapoor C, Yu E, Tarr TB, Titterton K, Zeng M, Park D, Niederst E, Wei D, Feng G, Boyden ES (2024) Multiplexed expansion revealing for imaging multiprotein nanostructures in healthy and diseased brain, Nature Communications 15(1):9722. (*, contributed equally)
Single-shot 20-fold expansion microscopy
Nature Methods | 2024Wang S*, Shin TW*, Yoder HB 2nd, McMillan RB, Su H, Liu Y, Zhang C, Leung KS, Yin P, Kiessling LL**, Boyden ES** (2024) Single-shot 20-fold expansion microscopy, Nature Methods, doi:10.1038/s41592-024-02454-9. Online ahead of print. (*, equal contribution; **, co-corresponding authors)
One-step nanoscale expansion microscopy reveals individual protein shapes
Nature Biotechnology | 2024Shaib AH, Chouaib AA, Chowdhury R, Altendorf J, Mihaylov D, Zhang C, Krah D, Imani V, Spencer RKW, Georgiev SV, Mougios N, Monga M, Reshetniak S, Mimoso T, Chen H, Fatehbasharzad P, Crzan D, Saal KA, Alawieh MM, Alawar N, Eilts J, Kang J, Soleimani A, Müller M, Pape C, Alvarez L, Trenkwalder C, Mollenhauer B, Outeiro TF, Köster S, Preobraschenski J, Becherer U, Moser T, Boyden ES, Aricescu AR, Sauer M, Opazo F, Rizzoli SO (2024) One-step nanoscale expansion microscopy reveals individual protein shapes, Nature Biotechnology doi:10.1038/s41587-024-02431-9. Online ahead of print.
Expansion microscopy using a single anchor molecule for high-yield multiplexed imaging of proteins and RNAs
PLoS One | 2023Cui Y*, Yang G*, Goodwin DR, O'Flanagan CH, Sinha A, Zhang C, Kitko KE, Shin TW, Park D, Aparicio S; CRUK IMAXT Grand Challenge Consortium; Boyden ES (2023) Expansion microscopy using a single anchor molecule for high-yield multiplexed imaging of proteins and RNAs, PLoS One 18(9):e0291506. (*, contributed equally)
3D Domain Adaptive Instance Segmentation via Cyclic Segmentation GANs
IEEE Journal of Biomedical and Health Informatics | 2023Lauenburg L, Lin Z, Zhang R, Santos MD, Huang S, Arganda-Carreras I, Boyden ES, Pfister H, Wei D (2023) 3D Domain Adaptive Instance Segmentation via Cyclic Segmentation GANs, IEEE Journal of Biomedical and Health Informatics 27(8):4018-4027.
Photo-expansion microscopy enables super-resolution imaging of cells embedded in 3D hydrogels
Nature Materials | 2023Günay KA, Chang TL, Skillin NP, Rao VV, Macdougall LJ, Cutler AA, Silver JS, Brown TE, Zhang C, Yu CJ, Olwin BB, Boyden ES, Anseth KS (2023) Photo-expansion microscopy enables super-resolution imaging of cells embedded in 3D hydrogels, Nature Materials 22(6):777-785.
Revealing nanostructures in brain tissue via protein decrowding by iterative expansion microscopy
Nature Biomedical Engineering | 2022Deblina Sarkar*, Jinyoung Kang*, Asmamaw T. Wassie*, Margaret E. Schroeder, Zhuyu Peng, Tyler B. Tarr, Ai-Hui Tang, Emily D. Niederst, Jennie Z. Young, Hanquan Su, Demian Park, Peng Yin, Li-Huei Tsai**, Thomas A. Blanpied** & Edward S. Boyden** (2022) Revealing nanostructures in brain tissue via protein decrowding by iterative expansion microscopy, Nature Biomedical Engineering 6:1057–1073. (*, contributed equally; **, co-corresponding)
Toward a more accurate 3D atlas of C. elegans neurons
BMC Bioinformatics | 2022Skuhersky M, Wu T, Yemini E, Nejatbakhsh A, Boyden E, Tegmark M (2022) Toward a more accurate 3D atlas of C. elegans neurons, BMC Bioinformatics 23(1):195.
ExCel: Super-Resolution Imaging of C. elegans with Expansion Microscopy
Methods in Molecular Biology | 2022Yu CJ*, Orozco Cosio DM*, Boyden ES (2022) ExCel: Super-Resolution Imaging of C. elegans with Expansion Microscopy, Methods in Molecular Biology 2468:141-203. (* contributed equally)
Tetra-gel enables superior accuracy in combined super-resolution imaging and expansion microscopy
Scientific Reports | 2021Lee H, Yu CC, Boyden ES, Zhuang X, Kosuri P (2021) Tetra-gel enables superior accuracy in combined super-resolution imaging and expansion microscopy, Scientific Reports 11(1):16944.
A highly homogeneous polymer composed of tetrahedron-like monomers for high-isotropy expansion microscopy
Nature Nanotechnology | 2021Gao R*, Yu CJ*, Gao L, Piatkevich KD, Neve RL, Munro JB, Upadhyayula S, Boyden ES (2021) A highly homogeneous polymer composed of tetrahedron-like monomers for high-isotropy expansion microscopy, Nature Nanotechnology 16:698–707. (*, equal contribution)
Barcoded oligonucleotides ligated on RNA amplified for multiplexed and parallel in situ analyses
Nucleic Acids Research | 2021Liu S, Punthambaker S, Iyer EPR, Ferrante T, Goodwin D, Fürth D, Pawlowski AC, Jindal K, Tam JM, Mifflin L, Alon S, Sinha A, Wassie AT, Chen F, Cheng A, Willocq V, Meyer K, Ling KH, Camplisson CK, Kohman RE, Aach J, Lee JH, Yankner BA, Boyden ES, Church GM (2021) Barcoded oligonucleotides ligated on RNA amplified for multiplexed and parallel in situ analyses, Nucleic Acids Research 49(10):e58.
Expansion Sequencing: Spatially Precise In Situ Transcriptomics in Intact Biological Systems
Science | 2021Alon S*, Goodwin DR*, Sinha A*, Wassie AT*, Chen F*, Daugharthy ER**, Bando Y, Kajita A, Xue AG, Marrett K, Prior R, Cui Y, Payne AC, Yao CC, Suk HJ, Wang R, Yu CJ, Tillberg P, Reginato P, Pak N, Liu S, Punthambaker S, Iyer EPR, Kohman RE, Miller JA, Lein ES, Lako A, Cullen N, Rodig S, Helvie K, Abravanel DL, Wagle N, Johnson BE, Klughammer J, Slyper M, Waldman J, Jané-Valbuena J, Rozenblatt-Rosen O, Regev A; IMAXT Consortium, Church GM***+, Marblestone AH***, Boyden ES***+ (2021) Expansion Sequencing: Spatially Precise In Situ Transcriptomics in Intact Biological Systems, Science 371(6528):eaax2656. (* equal contribution, ** key contributions to early stages of project, *** equal contribution, +co-corresponding authors)
In situ genome sequencing resolves DNA sequence and structure in intact biological samples
Science | 2020Payne AC*, Chiang ZD*, Reginato PL*, Mangiameli SM, Murray EM, Yao CC, Markoulaki S, Earl AS, Labade AS, Jaenisch R, Church GM, Boyden ES**, Buenrostro JD**, Chen F** (2020) In situ genome sequencing resolves DNA sequence and structure in intact biological samples, Science 371(6532):eaay3446. (* equal contribution, **co-corresponding)
Light microscopy based approach for mapping connectivity with molecular specificity
Nature Communications | 2020Shen FY, Harrington MM, Walker LA, Cheng HPJ, Boyden ES, Cai D (2020) Light microscopy based approach for mapping connectivity with molecular specificity, Nature Communications 11(1):4632.
Expansion Microscopy for Beginners: Visualizing Microtubules in Expanded Cultured HeLa Cells
Current Protocols in Neuroscience | 2020Zhang C*, Kang JS*, Asano SM, Gao R, Boyden ES (2020) Expansion Microscopy for Beginners: Visualizing Microtubules in Expanded Cultured HeLa Cells, Current Protocols in Neuroscience 92(1):e96. (*, contributed equally)
Expansion microscopy of C. elegans
eLife | 2020Yu CJ, Barry NC, Wassie AT, Sinha A, Bhattacharya A, Asano S, Zhang C, Chen F, Hobert O, Goodman MB, Haspel G, Boyden ES (2020) Expansion microscopy of C. elegans, eLife 9:e46249.
Expansion Microscopy of Lipid Membranes
bioRxiv | 2019Emmanouil D. Karagiannis*, Jeong Seuk Kang*, Tay Won Shin, Amauche Emenari, Shoh Asano, Leanne Lin, Emma K. Costa, IMAXT Grand Challenge Consortium, Adam H. Marblestone, Narayanan Kasthuri, Edward S. Boyden (2019) Expansion Microscopy of Lipid Membranes, bioRxiv 829903. (*, equal contribution)
Multiplexed and high-throughput neuronal fluorescence imaging with diffusible probes
Nature Communications | 2019Guo SM, Veneziano R, Gordonov S, Li L, Danielson E, Perez de Arce K, Park D, Kulesa AB, Wamhoff EC, Blainey PC, Boyden ES, Cottrell JR, Bathe M (2019) Multiplexed and high-throughput neuronal fluorescence imaging with diffusible probes, Nature Communications 10(1):4377.
Immuno-SABER enables highly multiplexed and amplified protein imaging in tissues
Nature Biotechnology | 2019Saka SK, Wang Y, Kishi JY, Zhu A, Zeng Y, Xie W, Kirli K, Yapp C, Cicconet M, Beliveau BJ, Lapan SW, Yin S, Lin M, Boyden ES, Kaeser PS, Pihan G, Church GM, Yin P (2019) Immuno-SABER enables highly multiplexed and amplified protein imaging in tissues, Nature Biotechnology 37(9):1080-1090.
A theoretical analysis of single molecule protein sequencing via weak binding spectra
PLoS One | 2019Rodriques SG, Marblestone AH, Boyden ES (2019) A theoretical analysis of single molecule protein sequencing via weak binding spectra, PLoS One 14(3):e0212868.
Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution
Science | 2019Ruixuan Gao*, Shoh M. Asano*, Srigokul Upadhyayula*, Igor Pisarev, Daniel E. Milkie, Tsung-Li Liu, Ved Singh, Austin Graves, Grace H. Huynh, Yongxin Zhao, John Bogovic, Jennifer Colonell, Carolyn M. Ott, Christopher Zugates, Susan Tappan, Alfredo Rodriguez, Kishore R. Mosaliganti, Shu-Hsien Sheu, H. Amalia Pasolli, Song Pang, C. Shan Xu, Sean G. Megason, Harald Hess, Jennifer Lippincott-Schwartz, Adam Hantman, Gerald M. Rubin, Tom Kirchhausen, Stephan Saalfeld, Yoshinori Aso, Edward S. Boyden**, Eric Betzig** (2019) Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution, Science 363(6424):eaau8302. (*, equal contribution, **, co-corresponding)
Expansion microscopy: principles and uses in biological research
Nature Methods | 2018Wassie AT*, Zhao Y*, Boyden ES (2019) Expansion microscopy: principles and uses in biological research, Nature Methods 16(1):33-41. (*, equal contributors)
Imaging cellular ultrastructures using expansion microscopy (U-ExM)
Nature Methods | 2018Gambarotto D, Zwettler FU, Le Guennec M, Schmidt-Cernohorska M, Fortun D, Borgers S, Heine J, Schloetel JG, Reuss M, Unser M, Boyden ES, Sauer M, Hamel V, Guichard P (2019) Imaging cellular ultrastructures using expansion microscopy (U-ExM), Nature Methods 16(1):71-74.
New observations in neuroscience using superresolution microscopy
Journal of Neuroscience | 2018Michihiro Igarashi, Motohiro Nozumi, Ling-Gang Wu, Francesca Cella Zanacchi, István Katona, László Barna, Pingyong Xu, Mingshu Zhang, Fudong Xue, Edward Boyden (2018) New observations in neuroscience using superresolution microscopy, Journal of Neuroscience 38 (44) 9459-9467.
Physical magnification of objects
Materials Horizons | 2018Boyden, E. S. (2018) Physical magnification of objects, Materials Horizons 6:11-13.
Expansion Microscopy: Protocols for Imaging Proteins and RNA in Cells and Tissues
Current Protocols in Cell Biology | 2018Asano SM*, Gao R*, Wassie AT*, Tillberg PW, Chen F, Boyden ES (2018) Expansion Microscopy: Protocols for Imaging Proteins and RNA in Cells and Tissues, Current Protocols in Cell Biology 80(1):e56. (*, co-first authors)
Expansion Microscopy: Enabling Single Cell Analysis In Intact Biological Systems
The FEBS Journal | 2018Alon, S.*, Huynh, G. H.*, Boyden, E. S. (2018) Expansion Microscopy: Enabling Single Cell Analysis In Intact Biological Systems, The FEBS Journal 286(8):1482-1494. (*, co-first authors)
Light sheet theta microscopy for rapid high-resolution imaging of large biological samples
BMC Biology | 2018Migliori B, Datta MS, Dupre C, Apak MC, Asano S, Gao R, Boyden ES, Hermanson O, Yuste R, Tomer R (2018) Light sheet theta microscopy for rapid high-resolution imaging of large biological samples, BMC Biology 16(1):57.
Expansion microscopy: development and neuroscience applications
Current Opinion in Neurobiology | 2018Karagiannis ED, Boyden ES (2018) Expansion microscopy: development and neuroscience applications, Current Opinion in Neurobiology 50:56-63.
Expansion Microscopy of Zebrafish for Neuroscience and Developmental Biology Studies
Proceedings of the National Academy of Sciences | 2017Freifeld L, Odstrcil I, Förster D, Ramirez A, Gagnon JA, Randlett O, Costa EK, Asano S, Celiker OT, Gao R, Martin-Alarcon DA, Reginato P, Dick C, Chen L, Schoppik D, Engert F, Baier H, Boyden ES (2017) Expansion microscopy of zebrafish for neuroscience and developmental biology studies, Proceedings of the National Academy of Sciences 114(50):E10799-E10808.
Glyoxal as an alternative fixative to formaldehyde in immunostaining and super-resolution microscopy
The EMBO Journal | 2017Richter KN, Revelo NH, Seitz KJ, Helm MS, Sarkar D, Saleeb RS, D'Este E, Eberle J, Wagner E, Vogl C, Lazaro DF, Richter F, Coy-Vergara J, Coceano G, Boyden ES, Duncan RR, Hell SW, Lauterbach MA, Lehnart SE, Moser T, Outeiro T, Rehling P, Schwappach B, Testa I, Zapiec B, Rizzoli SO (2017) Glyoxal as an alternative fixative to formaldehyde in immunostaining and super-resolution microscopy, The EMBO Journal 37(1):139-159.
Feasibility of 3D Reconstruction of Neural Morphology Using Expansion Microscopy and Barcode-Guided Agglomeration
Frontiers in Computational Neuroscience | 2017Yoon YG, Dai P, Wohlwend J, Chang JB, Marblestone AH, Boyden ES (2017) Feasibility of 3D Reconstruction of Neural Morphology Using Expansion Microscopy and Barcode-Guided Agglomeration, Frontiers in Computational Neuroscience 11:97.
Rapid Sequential in Situ Multiplexing with DNA Exchange Imaging in Neuronal Cells and Tissues
Nano Letters | 2017Wang Y, Woehrstein JB, Donoghue N, Dai M, Avendaño MS, Schackmann RCJ, Zoeller JJ, Wang SSH, Tillberg PW, Park D, Lapan SW, Boyden ES, Brugge JS, Kaeser PS, Church GM, Agasti SS, Jungmann R, Yin P (2017) Rapid Sequential in Situ Multiplexing with DNA Exchange Imaging in Neuronal Cells and Tissues, Nano Letters 17(10):6131-6139.
Q&A: Expansion microscopy
BMC Biology | 2017Gao R.*, Asano S.M.*, Boyden E.S. (2017) Q&A: Expansion microscopy, BMC Biology 15:50. (*, co-first authors)
Iterative expansion microscopy
Nature Methods | 2017Chang, J.-B., Chen, F., Yoon, Y.-G., Jung, E. E., Babcock H., Kang J.-S., Asano S., Suk H.-J., Pak N., Tillberg P.W., Wassie A., Cai D., Boyden E.S. (2017) Iterative expansion microscopy, Nature Methods 14:593-599.
Nanoscale imaging of RNA with expansion microscopy
Nature Methods | 2016Chen, F.*, Wassie, A.T.*, Cote, A.J., Sinha, A., Alon, S., Asano, S., Daugharthy, E.R., Chang, J.-B., Marblestone, A., Church, G.M., Raj, A., Boyden, E.S. (2016) Nanoscale Imaging of RNA with Expansion Microscopy, Nature Methods 13(8):679-84. (*, co-first authors)
Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies
Nature Biotechnology | 2016Tillberg, P.W.*, Chen, F.*, Piatkevich, K.D., Zhao, Y., Yu, C.-C., English, B.P., Gao, L., Martorell, A., Suk, H.-J., Yoshida, F., DeGennaro, E.M., Roossien, D.H., Gong, G., Seneviratne, U., Tannenbaum, S.R., Desimone, R., Cai, D., Boyden, E.S. (2016) Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies, Nature Biotechnology 34:987-992. (*, co-first authors)
Automated scalable segmentation of neurons from multispectral images
Advances in Neural Information Processing Systems | 2016Uygar Sümbül, Douglas Roossien, Dawen Cai, Fei Chen, Nicholas Barry, John P. Cunningham, Edward Boyden, Liam Paninski (2016) Automated scalable segmentation of neurons from multispectral images, Advances in Neural Information Processing Systems 29 (NIPS 2016).
Programmable RNA-binding protein composed of repeats of a single modular unit
Proceedings of the National Academy of Sciences | 2016Adamala, K.P.*, Martin-Alarcon, D. A.*, Boyden E. S. (2016) Programmable RNA-binding protein composed of repeats of a single modular unit, Proceedings of the National Academy of Sciences 113(19):E2579–E2588. (*, co-first authors)
Expansion Microscopy
Science | 2015Chen, F.*, Tillberg, P.W.*, Boyden, E.S. (2015) Expansion Microscopy, Science 347(6221):543-548. (*, equal contribution)