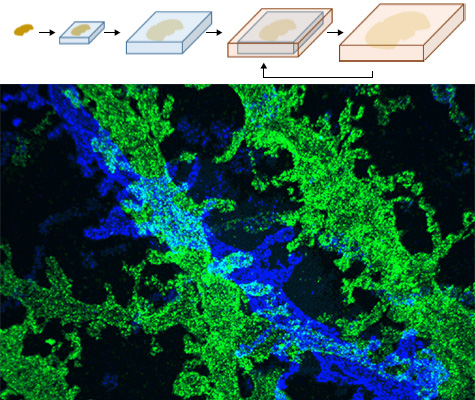
We recently developed a method called expansion microscopy, in which preserved biological specimens are physically magnified by embedding them in a densely crosslinked polyelectrolyte gel, anchoring key labels or biomolecules to the gel, mechanically homogenizing the specimen, and then swelling the gel–specimen composite by ~4.5× in linear dimension. Here we describe iterative expansion microscopy (iExM), in which a sample is expanded ~20×. After preliminary expansion a second swellable polymer mesh is formed in the space newly opened up by the first expansion, and the sample is expanded again. iExM expands biological specimens ~4.5 × 4.5, or ~20×, and enables ~25-nm-resolution imaging of cells and tissues on conventional microscopes. We used iExM to visualize synaptic proteins, as well as the detailed architecture of dendritic spines, in mouse brain circuitry.