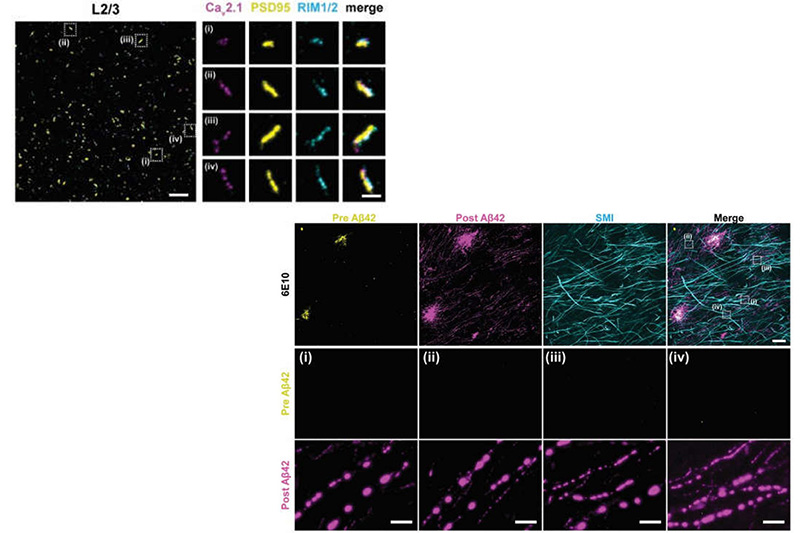
Many crowded biomolecular structures in cells and tissues are inaccessible to labelling antibodies. To understand how proteins within these structures are arranged with nanoscale precision therefore requires that these structures be decrowded before labelling. Here we show that an iterative variant of expansion microscopy (the permeation of cells and tissues by a swellable hydrogel followed by isotropic hydrogel expansion, to allow for enhanced imaging resolution with ordinary microscopes) enables the imaging of nanostructures in expanded yet otherwise intact tissues at a resolution of about 20 nm. The method, which we named ‘expansion revealing’ and validated with DNA-probe-based super-resolution microscopy, involves gel-anchoring reagents and the embedding, expansion and re-embedding of the sample in homogeneous swellable hydrogels. Expansion revealing enabled us to use confocal microscopy to image the alignment of pre-synaptic calcium channels with post-synaptic scaffolding proteins in intact brain circuits, and to uncover periodic amyloid nanoclusters containing ion-channel proteins in brain tissue from a mouse model of Alzheimer’s disease. Expansion revealing will enable the further discovery of previously unseen nanostructures within cells and tissues.
Original biorxiv preprint: version 1 posted on August 30, 2020, and available [here]