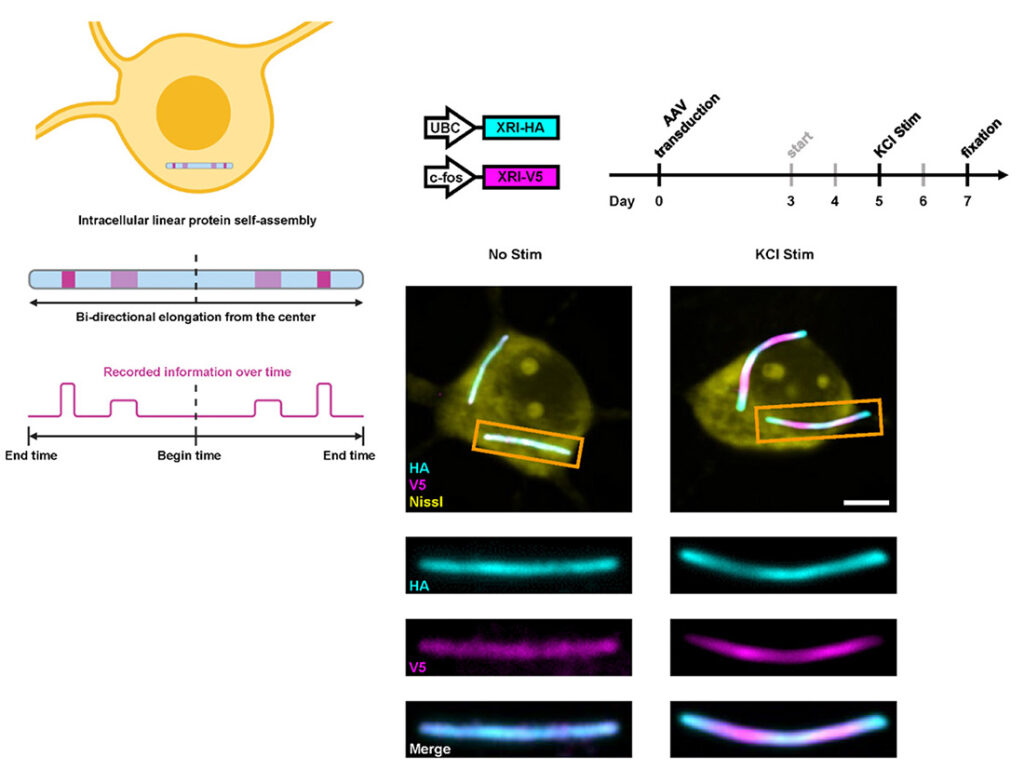
Observing cellular physiological histories is key to understanding normal and disease-related processes. Here we describe expression recording islands—a fully genetically encoded approach that enables both continual digital recording of biological information within cells and subsequent high-throughput readout in fixed cells. The information is stored in growing intracellular protein chains made of self-assembling subunits, human-designed filament-forming proteins bearing different epitope tags that each correspond to a different cellular state or function (for example, gene expression downstream of neural activity or pharmacological exposure), allowing the physiological history to be read out along the ordered subunits of protein chains with conventional optical microscopy. We use expression recording islands to record gene expression timecourse downstream of specific pharmacological and physiological stimuli in cultured neurons and in living mouse brain, with a time resolution of a fraction of a day, over periods of days to weeks.
Original biorxiv preprint: version 1 posted on October 13, 2021, and available [here]
